Artifacts annotations in anesthesia blood pressure data by man and machine
Abstract
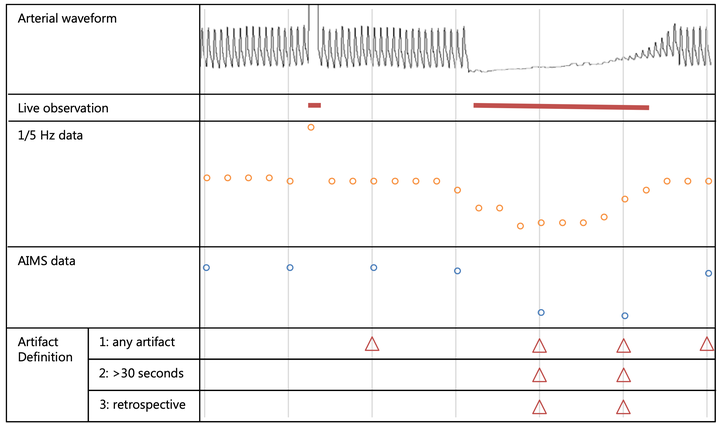
Physiologic data from anesthesia monitors are automatically captured. Yet erroneous data are stored in the process as well. While this is not interfering with clinical care, research can be affected. Researchers should find ways to remove artifacts. The aim of the present study was to compare different artifact annotation strategies, and to assess if a machine learning algorithm is able to accept or reject individual data points. Non-cardiac procedures requiring invasive blood pressure monitoring were eligible. Two trained research assistants observed procedures live for artifacts. The same procedures were also retrospectively annotated for artifacts by a different person. We compared the different ways of artifact identifications and modelled artifacts with three different learning algorithms (lasso restrictive logistic regression, neural network and support vector machine). In 88 surgical procedures including 5711 blood pressure data points, the live observed incidence of artifacts was 2.1% and the retrospective incidence was 2.2%. Comparing retrospective with live annotation revealed a sensitivity of 0.32 and specificity of 0.98. The performance of the learning algorithms which we applied ranged from poor (kappa 0.053) to moderate (kappa 0.651). Manual identification of artifacts yielded different incidences in different situations, which were not comparable. Artifact detection in physiologic data collected during anesthesia could be automated, but the performance of the learning algorithms in the present study remained moderate. Future research should focus on optimization and finding ways to apply them with minimal manual work. The present study underlines the importance of an explicit definition for artifacts in database research.